Image processing workhorses
There are many software packages for processing EM images. Because of their flexibility, I do most of my image processing with SPIDER and SPARX/EMAN2.
Many routine tasks are automated in my SPIDER batch procedures. Simply copy the batch procedure to your project directory, modify the input parameters, and run.
My SPIDER batch procedures rely on a library of SPIDER scripts. These can be also be called interactively.
For improved image format compatibility and to take advantage of the extensive capabilities of python, I have begun rewriting my SPIDER scripts to run using SPARX/EMAN2. Currently this has only been implemented for particle windowing and preprocessing. For 3D refinement and 2D alignment and clustering SPARX has fairly straightforward command-line programs (sxali3d.py and sxisac.py).
Manipulating and viewing images and volumes
Converting images
Scanned micrographs are often in TIFF format and can be converted with Bsoft's "bimg" command or Spider's "CP FROM RAW" command. EMAN's "proc2d" and EMAN2's "e2proc2d" also convert TIFF images to SPIDER format but will be mirrored top-bottom. I think Imagic's "em2em" also mirrors TIFF-to-SPIDER conversion.
CCD frames are typically in MRC format and can be converted with "bimg", "proc2d", "e2proc2d", or Spider's "CP FROM CCP4" command. Spider's "CP FROM MRC" command fails on MRC images (in version 13). Conversion can also be done with "em2em" but will mirror your images.
Volume display
In UCSF Chimera, SPIDER, and Web positive rotations are left-handed with thumb pointing in positive direction along axis of rotation. In Chimera volumes are viewed up the z-axis. To get the view that corresponds with the 0,0,0 projection (down the z-axis), just type open the volume and enter the Chimera command "turn x 180". From this position, you can find the view that corresponds to any projection by typing "turn z [phi]; turn y [theta]; turn z [psi]".
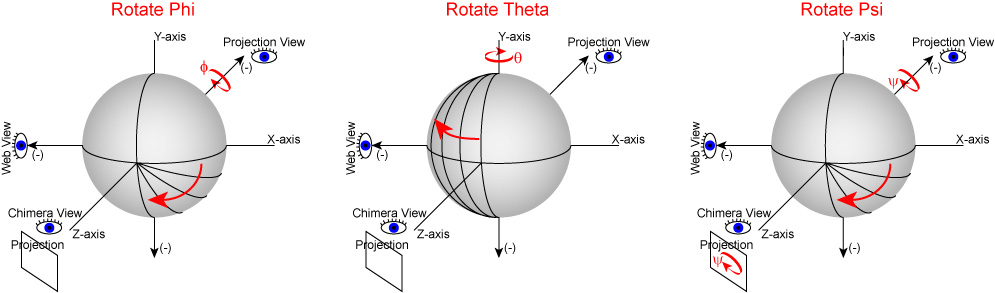
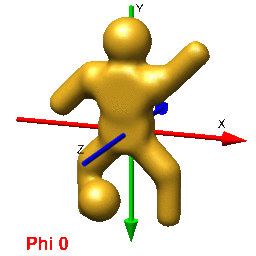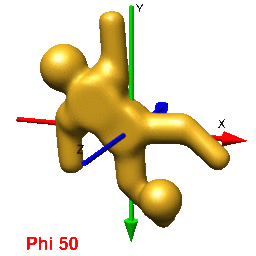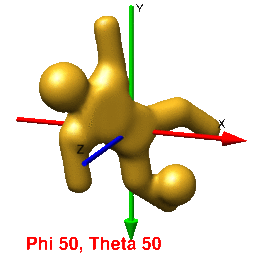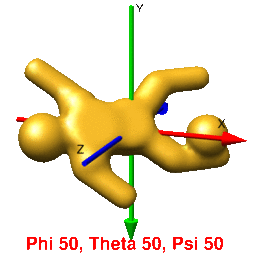
In Web's "Surface" volumes are viewed up the x-axis from negative[near] to positive[far], the y-axis is horizontal (negative[left] to positive[right]), and the z-axis is vertical (negative[up] to positive[down]) (i.e., right handed coordinate system). This is perpendicular and sideways to projection images generated by the SPIDER's "PJ 3Q" or "PJ SHAD" commands. After finding an orientation for your volume using Web's "Surface" to get the corresopnding view of your 3D-volume or 2D-projection you must (pre/post?)transform your Euler angles (phi (z), theta (y), psi (z)) by 0,90,90 using the "SA E" command. The resulting angles can then be used with the "RT 3D" or "PJ 3Q" commands to get the intended orientation or projection of the volume.
To do the reverse operation, as in the situation where you know the projection Euler angles but want the same view with Web's Surface, use the "SA E" command to (pre/post?)transform those angles (phi (z), theta (y), psi (z)) by 90,90,180.
When converting a pdb file into a Spider volume with the "CP FROM PDB" command, to preserve orientation it must be rotated by 0,180,90. My @pdb2spi script will take care of these operations for you.
Converting SPIDER volumes from CCP4
Since Spider's "CP TO CCP4" command fails (in version 13) I recommend using Imagic's "em2em". The problem is that "em2em" rotates a volume by 180,180,0 (phi, theta, psi) due to mirroring twice. So first use the "RT 3D" command to rotate the volume by 0,-180,-180 before converting with "em2em". This is useful for fitting pdb structures into the resulting ccp4 formatted volumes and having the fitted pdb coordinates match the original Spider volume.
Alignment of two volumes
The simplest way to match volumes or fit pdb's into volumes is using UCSF Chimera's "fit" command. This operation can even iteratively fit multiple maps or pdb's. Pdb coordinates can be converted to maps on the fly with the "molmap" command. Volume subtraction can be done with the "vop" operation.
To automate the fitting Spider's "OR 3Q" aligns two volumes but sticks in local minima. It is better to use a projection matching procedure to generate a close match between two volumes prior to using this command.
Alternatively, you can use CoAn (N.Volkmann) to align volumes with the maptran (matches bounding box), mapgrid (scales boxes), and fitmap (aligns volumes). Typically then to subtract one volume from the other, use the mapdif command.
I think Situs "colacor" command may also fit two volumes, but I have not used this.