Spider Batch Procedures
For a description of these procedures:
Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis.
Nat Struct Mol Biol. 2009 Feb;16(2):190-7.
Brignole EJ, Asturias FJ. Single-particle electron microscopy of animal fatty acid synthase describing macromolecular rearrangements that enable catalysis.
Methods in Enzymology. 2010 483:179-202.
If these procedures are useful to you, then please cite these references as appropriate for your application.
To run these batch procedures:
1) Download EdSpiderLibrary.tar and untar:
tar xvf EdSpiderLibrary.tar
A directory called ./spiproc/ will be created.
The ./spiproc/bat/ folder contains the batch procedures (described below).
The ./spiproc/lib/ directory contains the
library of scripts
that are modular utilities used by the batch procedures.
2) Set the SPPROC_DIR environment variable in your .cshrc so that the batch procedures can find the scripts library:
setenv SPPROC_DIR /path/to/spiproc/lib/
This also makes it easy to call the scripts from an interactive Spider session. For example:
OPERATION: @listfiles
3) Test the batch procedures by running ./spiproc/TestSpider.csh.
Before running this shell script you must edit the path to your spider executable at the top of TestSpider.csh.
This script will generate a mock tilt-pair dataset and process the particles to give you RCT volumes.
You can use this opportunity to familiarize yourself with the directory organization and of a typical project
and the files output by each batch procedure.
4) To run a batch procedure, copy the batch procedure from the spiproc/bat directory to your project directory, edit the parameters in the header, and execute.
For example:
cp /home/brignole/spiproc/bat/RandomConicalTilt.spi /my/project/bat/rct1.spi
cd /my/project/
vi bat/rct1.spi
spider spi @bat/rct1
Tip: To run a spider script that continues after logging off (this for 64bit machines):
nohup /fa/4Dem/spider_17.12/spider/bin/spider_linux_mpfftw_intel64
spi/dat @procedurename > outfile.txt &
Batch procedures are typically run in this order:
1) Assess Defocus/Drift/Astigmatism
PowerspecDefocus.spi - Generates power spectra and calculates defocus for each micro
2) Window Particles
WindowTiltPairs.spi - Windows raw particles from tilt pair images
WindowParticles.spi - Windows raw particles from single images
Tip: make particle box about 50% larger than particle diameter.
3) Preprocess Particles
PreprocessParticles.spi - Interpolate, Crop, Normalize, Filter, and Mask corners of a raw particle stack
Tip: make high pass filter about 40% larger than particle diameter and low pass about 18A is fine for neg stain.
4) Reference-Free Alignment
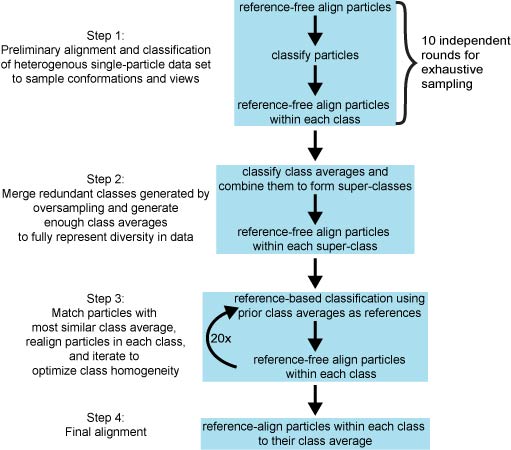
FreeAlign.spi - Performs 10 independent free-alignments, then aligns results and parameters from each round. (see Step 1 below)
5) Classification
MakeClasses.spi - Classifies particles from each alignment round then free-align particles in each class and align averages. (see Step 1 in Figure below)
MergeClasses.spi - Merge classes from each alignment round by classification of averages. Then free-align particles in each superclass. (see Step 2 in Figure below)
QuickAlignClassify.spi - Reference-free alignment followed by rotationally invariant classification. Then free-align in each class. This generates quick & dirty class averages.
|
6) Iterative Reference Classification / Free Alignment IterativeClassifyAlign.spi - Iterates multi-ref classification and free-alignment using class averages as initial references. (see Steps 3 & 4 in Figure below) |

|
7) RCT Volume Reconstruction
RandomConicalTilt.spi - Back projects a volume for each class. Refines volumes. Calculates resolution.