» about
I'm a Ph.D. student in the MIT Machine Learning Group and at the Broad Institute, working on probabilistic methods for learning from single-cell studies.
I am co-advised by Tamara Broderick (MIT CSAIL) and Aviv Regev (Broad Institute of MIT & Harvard).
Before that, I was a Molecular Biology major at Pomona College, a Fulbright scholar at the Australian Centre for Ecogenomics, an intern at Fast Forward Labs, and a cheese microbiologist at Jasper Hill.
» projects
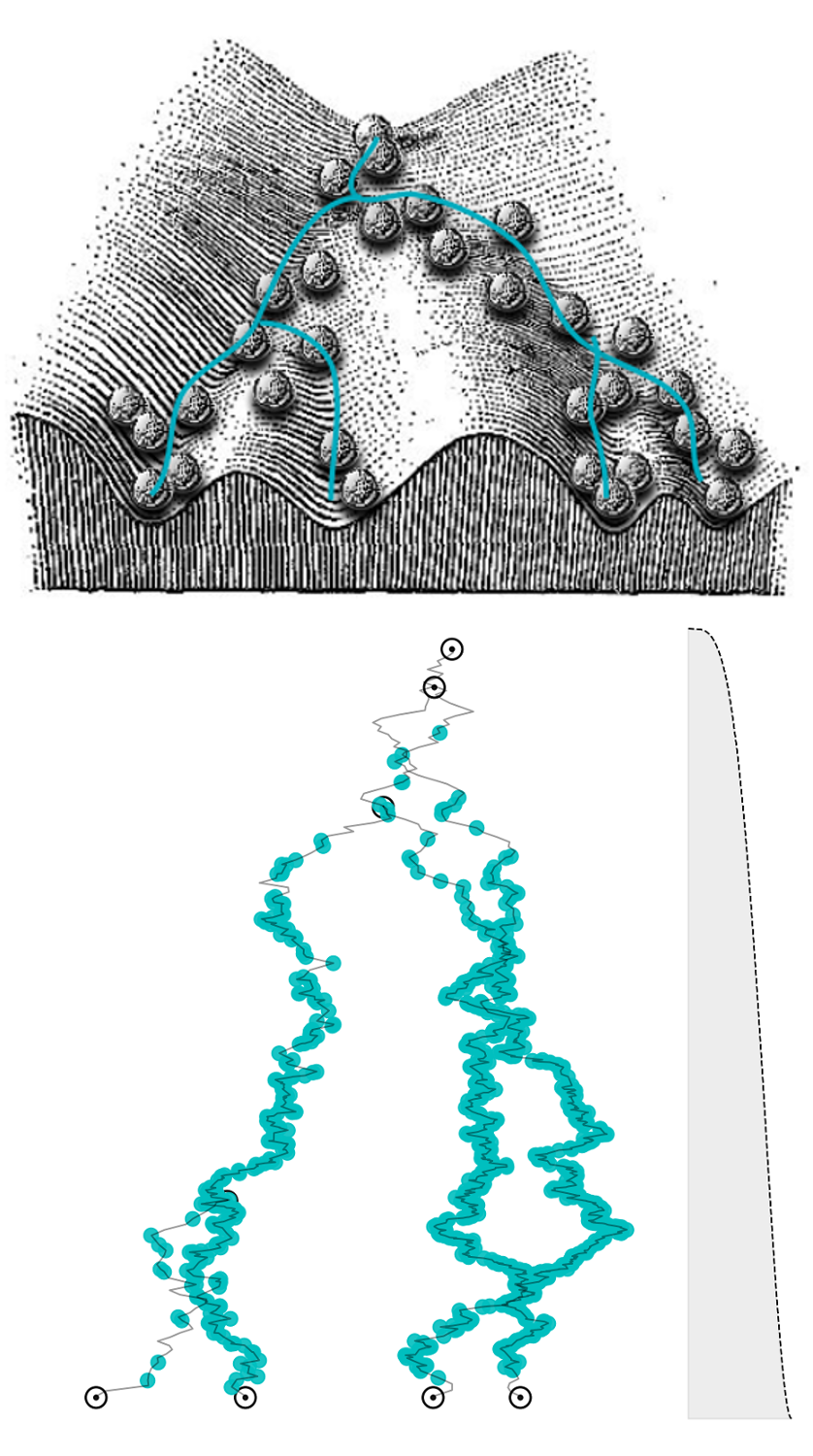
Bayesian tree models of cellular differentiation
[ MIT CSAIL, Broad Institute ]
Inferring latent cell state dynamics (along a probabilistic branching continuum) from static snapshots (single-cell profiles of gene expression).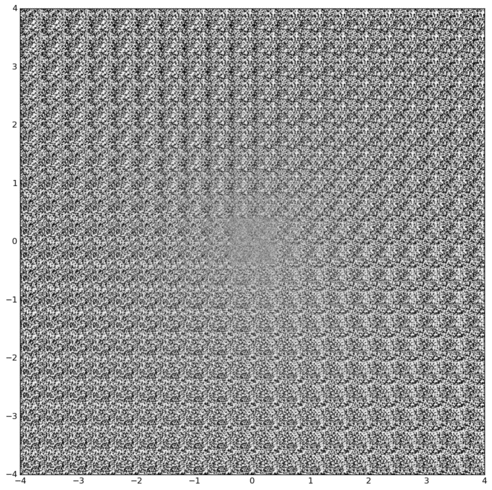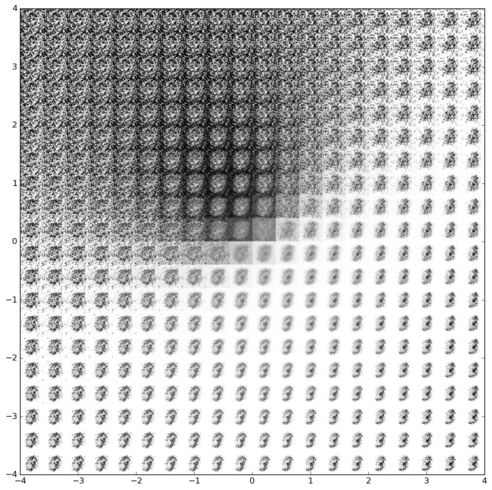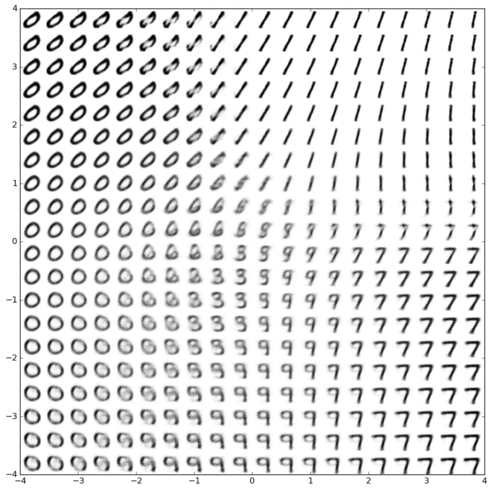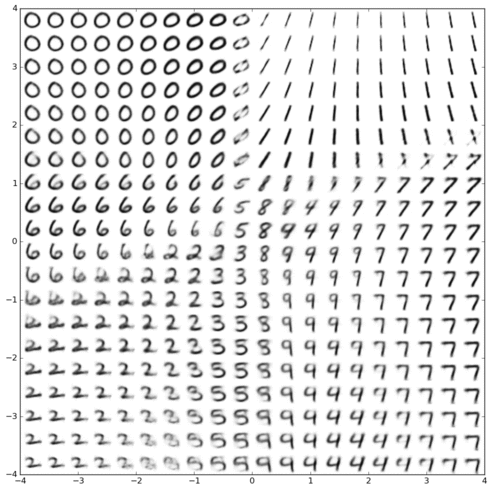
Variational autoencoders, in prose and code
[ Fast Forward Labs ]
Whitepaper-style blog series for FFL.
Microbial ecology and evolution
[ Australian Centre for Ecogenomics, Jasper Hill Farm, Harvard Systems Biology ]
Computational and experimental approaches to microbial communities in marsupial guts, the human microbiome, and cheese.-
Shiffman M, RM Soo, PG Dennis, M Morrison, GW
Tyson, P Hugenholtz (2017) Gene and genome-centric
analyses of koala and wombat fecal microbiomes point to
metabolic specialization for Eucalyptus digestion.
PeerJ.
(paper, blogpost) - Shiffman M, J Button, RJ Dutton (2014) Cheese as a model microbial ecosystem. International Society for Microbial Ecology (ISME) conference. (poster)
- Shiffman M, LM Seligman (2013) Evolutionary dynamics of intein-encoded homing endonucleases. (thesis, figs)
- Shiffman M, BM Charalambous (2012) The search for archaeal pathogens. Reviews in Medical Microbiology. (paper)
