Visualizing Timelapses and Videos with 3D Slicer
A quick tutorial - contact: Adrian Dalca
3D Slicer is a powerful open-source software package for 3D medical image analysis and visualization, developed at SPL, BWH. We use it very often in our medical work. Due to its intrinsic property of neatly dealing with 3D medical images, giving very nice methods for visualization and evaluation of such data, I thought Slicer would give a unique way of looking at video timelapses. This page includes some notes on how to get started with this process using Slicer (and optionally Matlab). If you use Slicer and find it useful, please cite the original authors. Slicer works on all major platforms.
Getting Started
- First, Download Slicer from SPL. The instructions on this tutorial were tested with Slicer version 4.1, but all stable versions should work.
- If you have your frames into individual images and are nicely numbered:
- you can simply load them into slicer by selecting File->Add data-->choose files to add, and selecting the first frame image. Then make sure you select "Show options" and unselect 'Single File'. This should load the entire timelapse. You can save the entire volume as one file if you wish.
- Download the NIfTI and ANALYZE MATLAB toolbox by Jimmy Shen (BSD License). Add it to your path in Matlab (>> help addpath). Besides giving us tools to build and save nifti files, the toolbox allows for quick and simple visualization and processing. It's a neat tool to have if you wish to process your files in MATLAB through these formats.
- Download some matlab functions I wrote for getting your files into slicer.
Many thanks to Ramesh Sridharan for correct outputting of RGB channels and options.- video2nii.m transforms your video file into a NIfTI stucture (and optionally a file) which you can then open in slicer. A NIfTI file (file.nii or file.nii.gz) is a convenient volume file that includes the entire timelapse and can be read into slicer.
- frames2nii.m to gather individual images (frames of a timelapse) into a NIfTI structure/file.
- save_niigz.m to save gzipped version of NIfTI file.
- load_niigz.m to load gzipped version of NIfTI file.
- run video2nii or frames2nii, which gives you a nii.gz file. You can process the nii struct further into matlab, or just load it into slicer using File->Add Volume (no extra options are necessary).
Visualization and processing in 3D Slicer.
There are many modules which can be of great use. Below are some ideas and screenshots/screenvids.
- Simple Visualization in 2D different coordinates (XY or XT) and 3D:
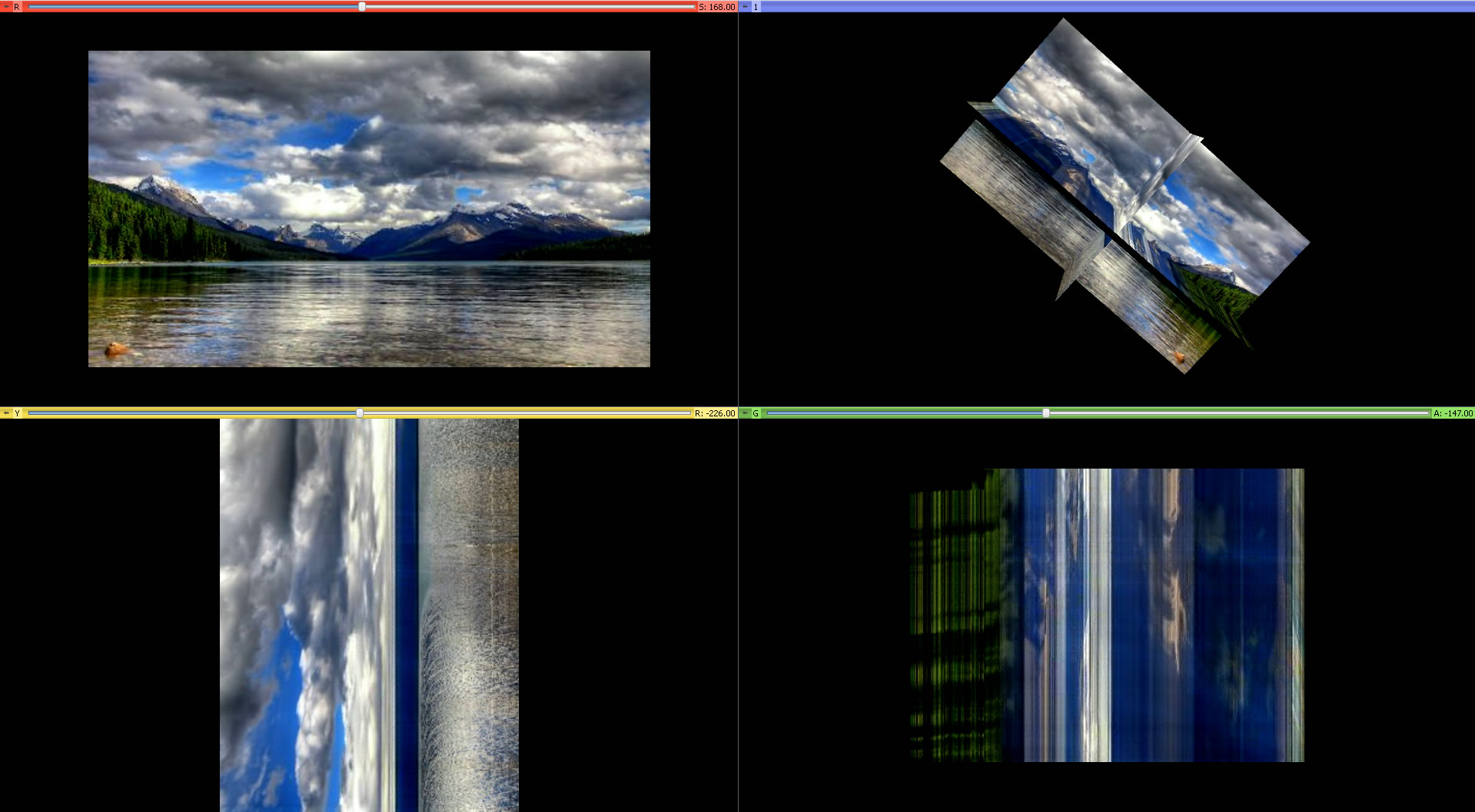
- Observing irregular motion in time plane. Below, the white irregular paths in the XT plane are boats moving on the river. The XT plane crossing the normal XY plane is chosen at the level where the river meets the mountains.

- Slicer allows for easy (linear) refactoring of slices in the volume. In the image below, the red outline is a slice that's diagonal through the XYT (spacetime) volume. The top left are clouds at the end of the timelapse, the bottom right are waves at the beginning of the timelapse.
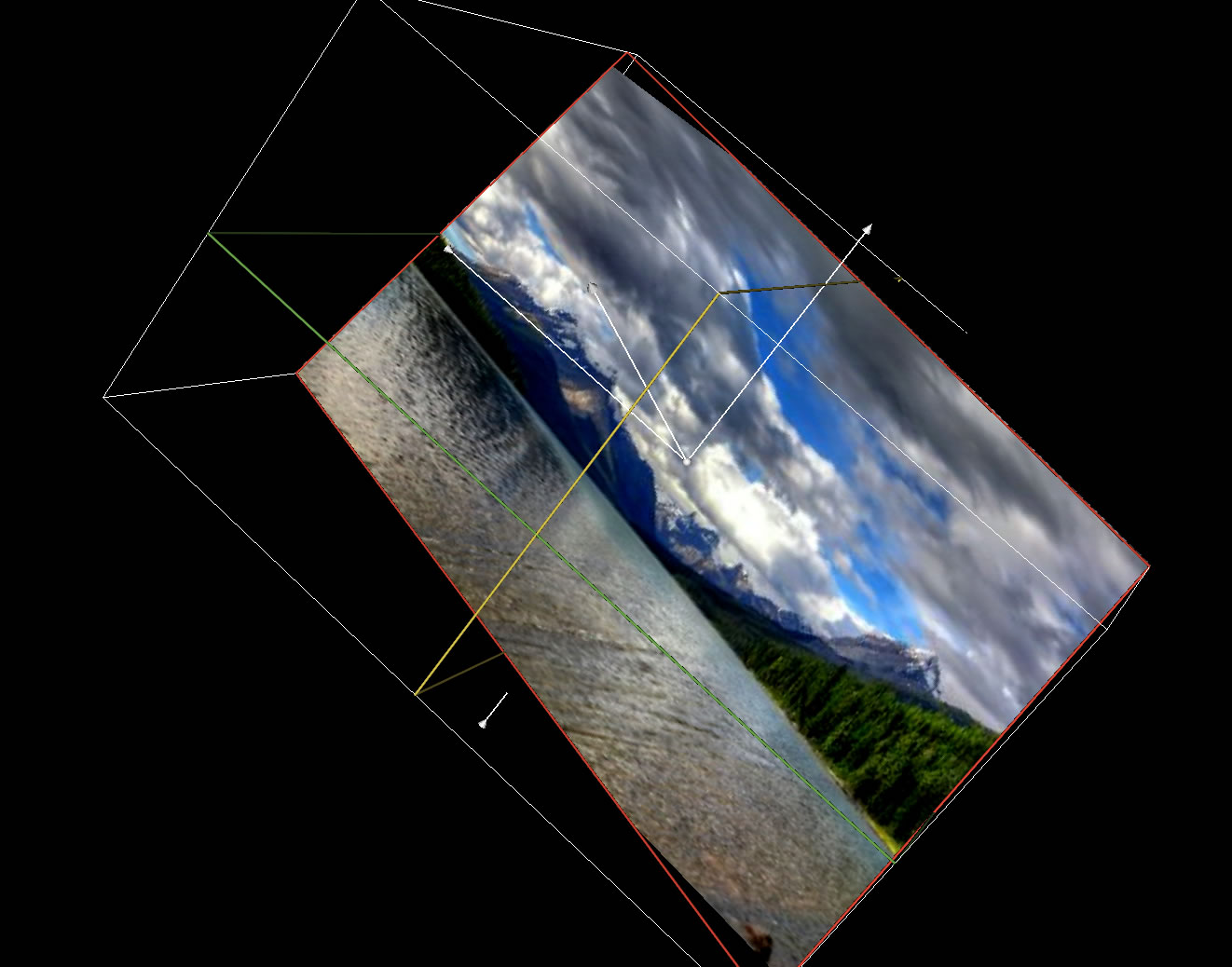
- By moving a view slider, one can see the timelapse flow along various dimensions.
Trivially, you can see the change of your scene with respect to time (as it would show in a video), but also see the change in any new coordinate frame.
- Slicer allows many other features through its modules, although they are tuned to medical data. You could, for example, obtain 3D (space-time) segmentations, and create 3D models of these to visualize their share and behaviour in space-time.