Assistant Professor in Computer Science, UIUC
Ph.D. in Computer Science, MIT
Ex Post-doc at Baker Lab, Institute for Protein Design, UW
Email:geliu@illinois.edu, geliu@csail.mit.edu
[
Google scholar]
[
Github]
[
CV]
I recently joined the Department of Computer Science of the University of Illinois at Urbana-Champaign (UIUC) as an Assistant Professor. I am looking for PhD students starting in 2025 Fall (application due Dec 2024). I received my PhD from MIT EECS department, advised by professor David Gifford from Computer Science and Artificial Intelligence Laboratory (CSAIL).Prior to joining UIUC, I spent an amazing year at the Institute for Protein Design as a postdoctoral researcher advised by professor David Baker. My research develops scalable, reliable, and geometry-aware deep generative models and sequential optimization techniques, as well as novel experiment frameworks and computational tools, for solving important problems in synthetic biology, immunology, and molecular biology, that go beyond just predictive modeling. I am especially interested in geometric DL and multi-modal flow matching & diffusion methods for computational design of functional and therapeutic molecules, including but not limited to antibody design, enzyme and catalyst design and vaccine design. Machine learning wise I'm working on geometry-aware multimodal flow matching/diffusion models, sequential optimization and RL with generative priors, post-training and steering foundational generative models, active learning and model uncertainty. My PhD thesis won the MIT EECS George M. Sprowls Ph.D. Thesis Award in AI and Decision-Making in 2021.
I recieved my bachelor degree from Tsinghua University EE department, where I worked as a research assistant in Machine Learning and Computational Biology Group (IIIS), advised by Prof. Jianyang Zeng. I was a visiting scholar at CMU in 2014 summer and worked in Murphy Lab, Lane Center for Computational Biology, advised by professor Robert F. Murphy.
For Prospective Students and Interns: I am actively looking for highly-motivated Ph.D. students and interns to work on problems in the exciting frontiers of ML and AI4Science. If you are interested in joining my group, please send me an email at
geliu[at]illinois[dot]edu with your CV and research summary. Students with strong motivation to do good science and make an impact in biomedicine while innovating in ML/CS are highly welcomed. Some of my research topics include but not limited to:
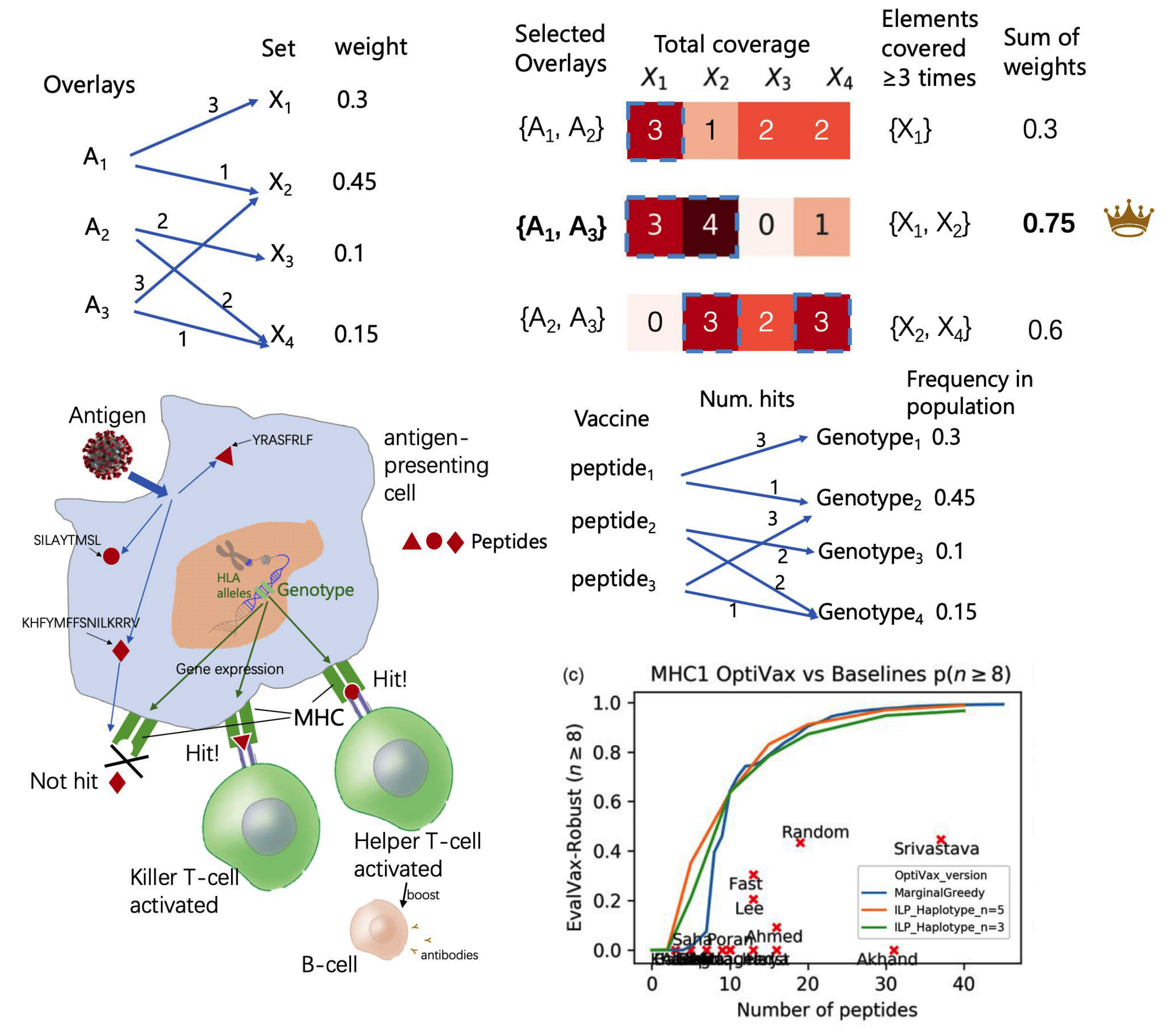
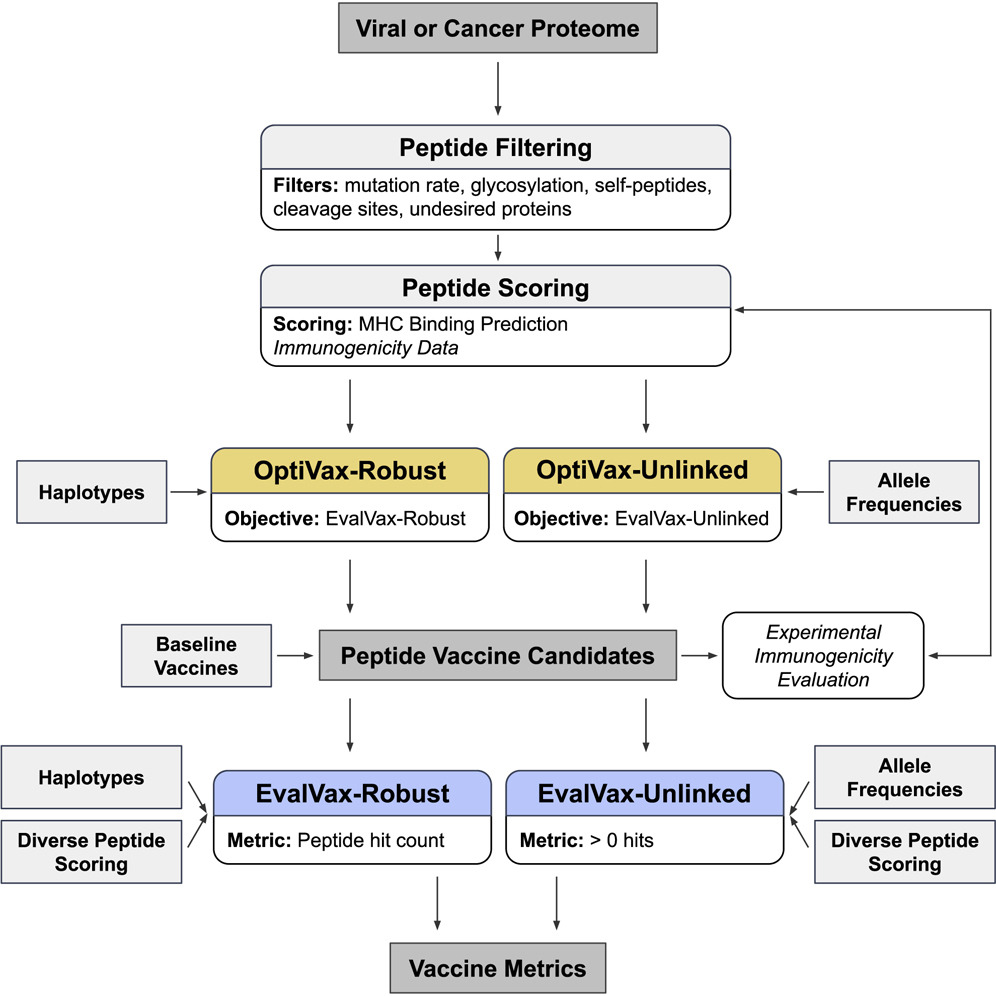
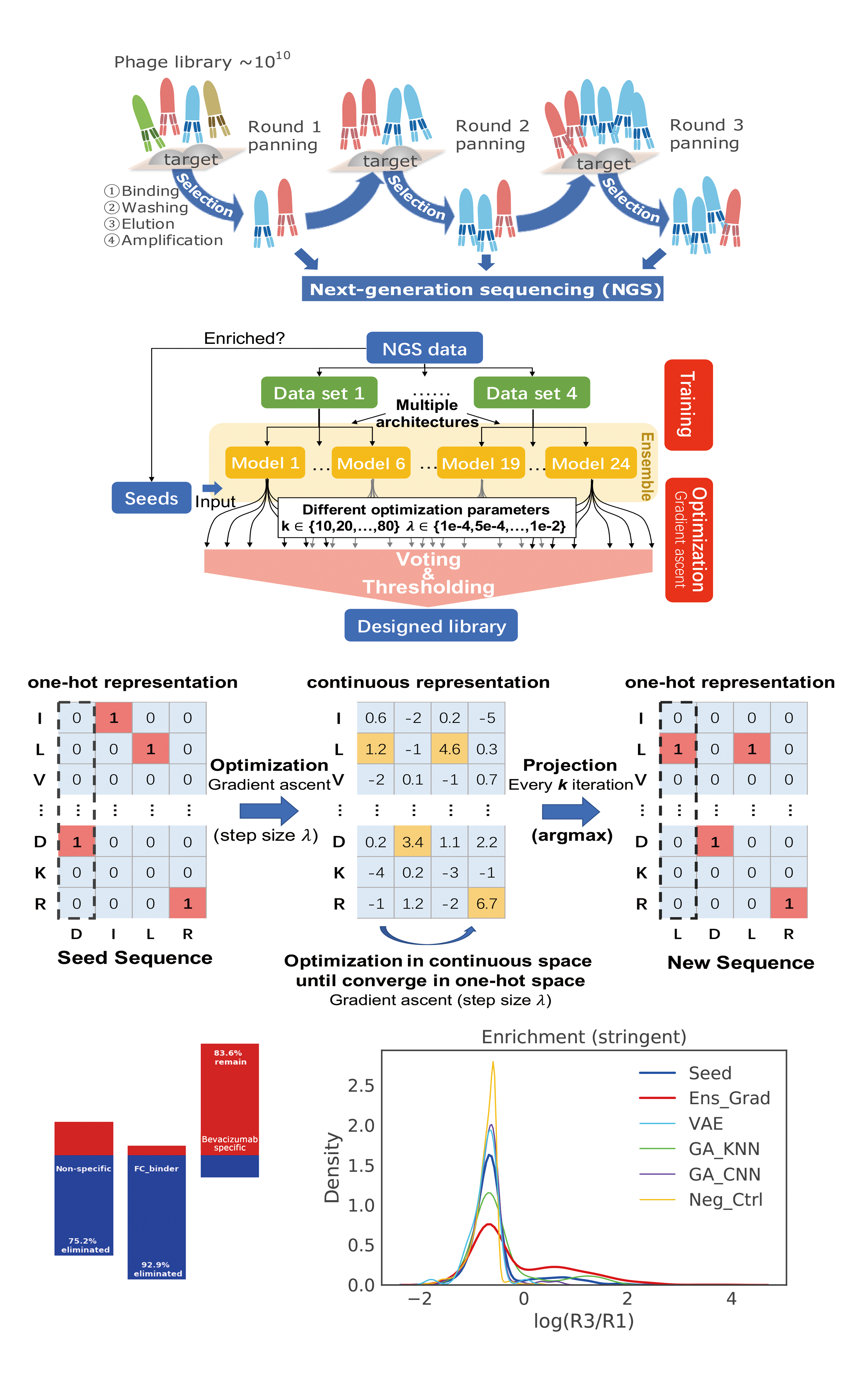
- Deep generative models and its application to functional biological molecule design (antibodies, peptide vaccines, enzymes) and drug discovery. We're particularly interesting in multimodal flow matching on manifolds and geometry-aware, physics-informed deep generative modeling, and solving complex guided and conditional generation tasks and inverse problems with diffusion/FM prior.
- Iterative experiment design and post-training with generative priors, using uncertainty-aware learning, online learning algorithms (e.g. RLHF, bandits), active learning, Bayesian optimization.
- Optimization: efficient algorithms for solving combinatorial optimization, discrete optimization, and black-box optimization problems in real-world biomedicine design problems.
 [Full image]
[Full image]

 [Full image]
[Full image]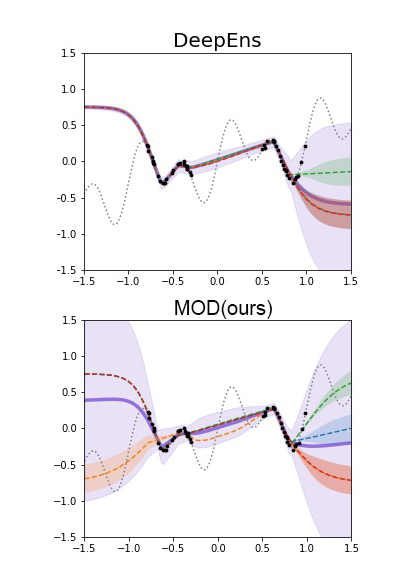
 [Full image]
[Full image] [Full image]
[Full image] [Full image]
[Full image]