|
I am a Senior Researcher at Microsoft Research in Cambridge, MA. I completed my PhD in Biophysics at Harvard University, where I worked with Sangeeta Bhatia at the Koch Institute for Integrative Cancer Research and was supported by the NSF Graduate Research Fellowship.
I received my Bachelor of Science in Computer Science and Molecular Biology from MIT,
where I conducted research with Tim Lu and was recognized as a Henry Ford II Scholar
and with the AMITA Senior Academic Award.
Email / CV / Google Scholar / Twitter |

|
|
|
My research focuses on engineering new technologies for precision medicine. I have developed new methods in machine learning and statistics, bioengineering, and nanotechnology and leveraged these approaches to create new diagnostic and therapeutic biotechnologies and to achieve new insights into cancer. Formerly Ava Soleimany. *Denotes co-first authorship |
|
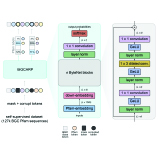
Carolina Rios-Martinez, Nicholas Bhattacharya, Ava P. Amini, Lorin Crawford, Kevin K. Yang bioRxiv, 2022 We develop a self-supervised masked language model for biosynthetic gene clusters in bacteria, and leverage it for natural product classification. |
 
|
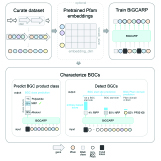
Ava P. Soleimany*, Carmen Martin-Alonso*, Melodi Anahtar*, Cathy S. Wang, Sangeeta N. Bhatia ACS Omega, 2022 pdf / code We build Protease Activity Analysis (PAA), a Python software package with a collection of data analytic and machine learning tools for analyzing protease activity data. |
|
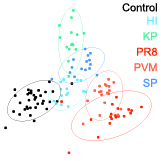
Melodi Anahtar, Leslie W. Chan, Henry Ko, Aditya Rao, Ava P. Soleimany, Purvesh Khatri, Sangeeta N. Bhatia PNAS, 2022 press / pdf / code We develop a sensor-based, ML-driven system to diagnose pneumonia and classify its etiology, using machine learning to classify directly from molecular barcodes. |
|
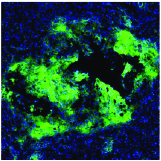
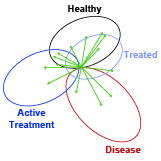
Jesse D. Kirkpatrick, Ava P. Soleimany, Jaideep S. Dudani, Heng-Jia Liu, Hilaire C. Lam, Carmen Priolo, Elizabeth P. Henske, Sangeeta N. Bhatia European Respiratory Journal, 2022 We establish a sensor-based, ML-driven diagnostic for noninvasive, real-time monitoring of disease in a preclinical model of lymphangioleiomyomatosis (LAM), a rare lung disease. |
|
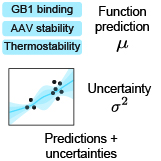
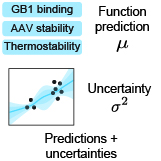
Kevin P. Greenman, Ava P. Soleimany, Kevin K. Yang ICLR Workshop on Machine Learning for Drug Discovery, 2022 We assess deep learning-based uncertainty quantification methods on protein sequence-function prediction tasks. |
|
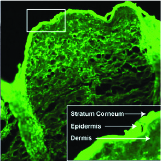
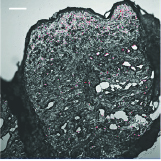
Ahmet Bekdemir, Eden E.L. Tanner, Jesse D. Kirkpatrick, Ava P. Soleimany, Samir Mitragotri, Sangeeta N. Bhatia Advanced Healthcare Materials, 2022 pdf / supplement We design an easily applicable, non-invasive formulation to deliver diagnostic nanosensors through the skin, enabling a sustained release diagnostic monitoring system for detecting thrombosis. |
|
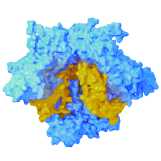
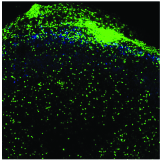
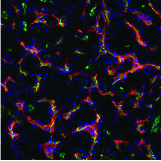
Aereas Aung, Ang Cui, Ava P. Soleimany, Maurice Bukenya, Heya Lee, Christopher A. Cottrell, Murillo Silva, Jesse D. Kirkpatrick, Parastoo Amlashi, Tanaka Remba, Shuhao Xiao, Leah Michelle Froehle, Wuhbet Abraham, Josetta Adams, Heikyung Suh, Phillip Huyett, Douglas S. Kwon, Nir Hacohen, William R. Schief, Sangeeta N. Bhatia, Darrell J. Irvine bioRxiv preprint, 2021 pdf / supplement We discover that protease activity and antigen breakdown are spatially heterogenous in lymph nodes, and that this spatially-compartmentalized antigen proteolysis can be exploited to enhance vaccine-induced antibody response. |
|
Ava P. Soleimany*, Jesse D. Kirkpatrick*, Cathy S. Wang, Alex M. Jaeger, Susan Su, Santiago Naranjo, Qian Zhong, Christina M. Cabana, Tyler Jacks, Sangeeta N. Bhatia bioRxiv preprint, 2021 pdf / supplement We engineer an integrated set of methods for measuring specific enzyme activities across the organismal, tissue, and cellular scales, and unify these methods into a methodological hierarchy that powers new biological insights into cancer. |
|
Jiang He*, Lior Nissim*, Ava P. Soleimany*, Adina Binder-Nissim, Heather E. Fleming, Timothy K. Lu, Sangeeta N. Bhatia ACS Synthetic Biology, 2021 pdf / supplement We design a sense-and-respond system that integrates a synthetic gene circuit and nanotechnology detection tools for tumor-specific expression of heterologous biomarkers. |
|
Ava P. Soleimany*, Alexander Amini*, Samuel Goldman*, Daniela Rus, Sangeeta N. Bhatia, Connor W. Coley ACS Central Science, 2021 pdf / supplement A fast, scalable approach for uncertainty quantification in neural networks enables uncertainty-aware molecular property prediction, accelerated property optimization, and guided virtual screening. |
|
Ava P. Soleimany*, Jesse D. Kirkpatrick*, Susan Su, Jaideep S. Dudani, Qian Zhong, Ahmet Bekdemir, Sangeeta N. Bhatia Cancer Research, 2021 pdf / supplement We engineer a new class of enzyme activity probes that can be applied to fresh-frozen tissue sections to spatially localize protease activty, enabling new insights into the biology of protease dysregulation. |
|
Alexander Amini, Wilko Schwarting, Ava Soleimany, Daniela Rus NeurIPS, 2020 pdf / press We develop a novel algorithm for fast, scalable uncertainty quantification in highly complex, non-linear neural networks trained for regression tasks. |
|
Naveen K. Mehta, Roma V. Pradhan, Ava P. Soleimany, Kelly D. Moynihan, Adrienne M. Rothschilds, Noor Momin, Kavya Rakhra, Jordi Mata-Fink, Sangeeta N. Bhatia, K. Dane Wittrup, Darrell J. Irvine Nature Biomedical Engineering, 2020 pdf / supplement / press We optimize the immunogenicity of peptide-based antitumor vaccines in mice by tuning their pharmacokinetics via fusion of the peptide epitopes to protein carriers. |
|
Ava P. Soleimany, Sangeeta N. Bhatia Trends in Molecular Medicine, 2020 Review detailing how integrating techniques from multiple disciplines has developed engineered diagnostics that are selectively activated in disease states, highlighting their potential to realize the goals of precision medicine. |
|
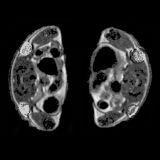
Jesse D. Kirkpatrick*, Andrew D. Warren*, Ava P. Soleimany*, Peter M. K. Westcott, Justin C. Voog, Carmen Martin-Alonso, Heather E. Fleming, Tuomas Tammela, Tyler Jacks, Sangeeta N. Bhatia Science Translational Medicine, 2020, *Co-first authors supplement / press / video We couple protease-responsive nanoparticle sensors with machine learning to engineer a sensitive and specific urinary test for lung cancer detection. |
|
Simone Schuerle, Maiko Furubayashi, Ava P. Soleimany, Tinotenda Gwisai, Wei Huang, Christopher A. Voigt, Sangeeta N. Bhatia ACS Synthetic Biology, 2020 supplement Magnetic nanoparticles that display genetically encoded targeting peptides to promote tumor accumulation and enhance MRI contrast. |
|
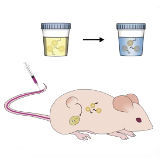
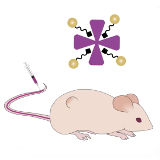
Colleen Loynachan*, Ava P. Soleimany*, Jaideep S. Dudani, Yiyang Lin, Adrian Najer, Ahmet Bekdemir, Qu Chen, Sangeeta N. Bhatia†, Molly M. Stevens† Nature Nanotechnology, 2019, *Co-first authors, †Co-corresponding authors supplement / data / press / video By leveraging the unique properties of catalytic nanomaterials, we develop a simple color-change urine test for detection of cancer in mice. |
|
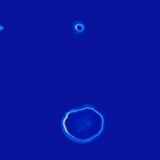
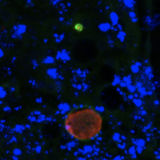
Ava P. Soleimany, Harini Suresh, Jose Javier Gonzalez Ortiz, Divya Shanmugam, Nil Gural, John Guttag, Sangeeta N. Bhatia ICML Workshop on Computational Biology, 2019 Convolutional neural networks for automated segmentation and uncertainty estimation of microscopy images of malaria infection. |
|
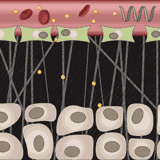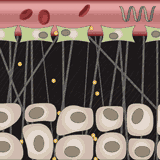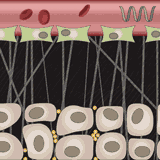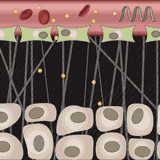
Simone Schuerle, Ava P. Soleimany, Tiffany Yeh, Giridhar M. Anand, Moritz Haberli, Heather E. Fleming, Nima Mirkhani, Famin Qiu, Sabine Hauert, Xiaopu Wang, Bradley J. Nelson, Sangeeta N. Bhatia Science Advances, 2019 supplement / press / video Engineered microrobots that use magnetism to push drug-delivery nanoparticles out of blood vessels and into diseased tissue. |

|
Alexander Amini*, Ava P. Soleimany*, Wilko Schwarting, Sangeeta N. Bhatia, Daniela Rus AAAI/ACM Conference on Artificial Intelligence, Ethics, and Society, 2019, *Co-first authors MIT press / VentureBeat press Generalizable algorithm for mitigating hidden biases within training data, by leveraging learned latent distributions to adaptively re-weight the importance of certain data points while training. |
|
Alexander Amini, Ava Soleimany, Sertac Karaman, Daniela Rus, NeurIPS Workshop on Bayesian Deep Learning, 2017 Estimating uncertainty in neural networks for end-to-end control by exploiting feature map correlations during training. |
 
|
Nathaniel Roquet, Ava P. Soleimany, Alyssa C. Ferris, Scott Aaronson, Timothy K. Lu Science, 2016 supplement / press / blog Programming biological state machines that enable cells to remember and respond to a series of events. |
|
In addition to research, I am passionate about education and leadership, and strive to help and empower others to excel in their own pursuits. |
 
|
I am an organizer and lecturer for Introduction to Deep Learning (6.S191), MIT’s official introductory course on deep learning foundations and applications. Together with Alexander Amini, I have organized and developed all aspects of the course, including developing the curriculum, teaching the lectures, creating software labs, and collaborating with industry sponsors. All materials can be found online on the course website. |
 |
Co-founder, Momentum AI
I am a co-founder and director for Momentum AI, an outreach program that teaches AI and machine learning to under-resourced and under-served high school
students from the greater Boston area. Our two-week capston program is a free, project-based deep dive into AI and is held on MIT's campus.
|
 |
Teaching Fellow, Harvard MCB294, Fall 2019, with Nancy Kleckner
|

|
Teaching Assistant, MIT 7.05, Spring 2016, with Matt Vander Heiden and Mike Yaffe
Teaching Assistant, MIT 7.05, Spring 2015, with Matt Vander Heiden and Mike Yaffe Captain, MIT Varsity Women's Tennis, 2014-2016 MIT Varsity Women's Tennis, 2012-2016 |