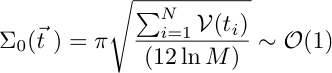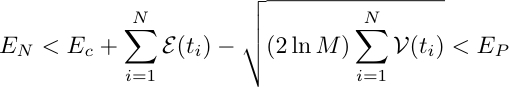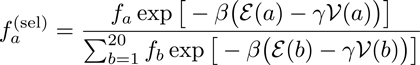Thymic Selection
![]() Immunological Tolerance of the T cell repertoire is shaped in the thymus gland:
Immunological Tolerance of the T cell repertoire is shaped in the thymus gland:
where a diverse repertoire of thymocytes (expressing distinct TCRs, produced by random recombination in bone marrow)
is culled by encounters with self-pMHC, via
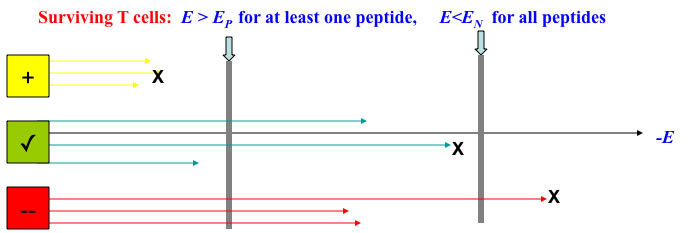
Positive selection: TCR must bind sufficiently strongly to at least one self-pMHC
(implicated in MHC restriction, and sensitivity).
Negative selection: TCR must not bind any self-pMHC too strongly
(deleting autoimmune TCR).
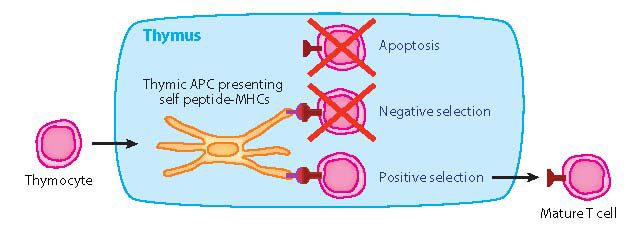
![]() Model for Thymic selection: [Kosmrlj, Jha, Huseby, Kardar, & Chakraborty, PNAS 105, 16671 (2008) (off-line)]
Model for Thymic selection: [Kosmrlj, Jha, Huseby, Kardar, & Chakraborty, PNAS 105, 16671 (2008) (off-line)]
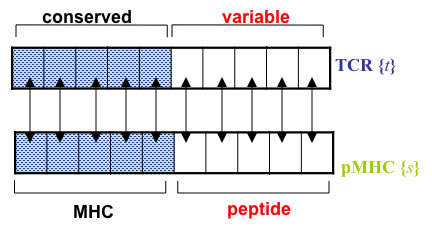
Many candidate TCR, modelled by sequences of N amino-acids, are generated randomly (ignoring any V(D)J generation bias).
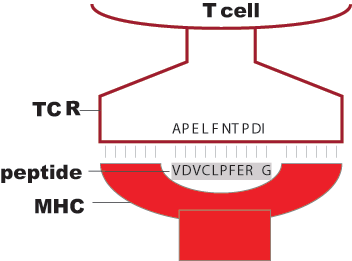
Binding energy E: Pairwise interactions between aligned amino-acids of peptide and TCR (Miazawa-Jernigan matrix J)
The binding energy of any TCR is calculated against a repertoire of M self-peptides.
Binding energy E: Pairwise interactions between aligned amino-acids of peptide and TCR (Miazawa-Jernigan matrix J)
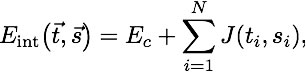
What are characteristics of amino-acids in the post-selection TCR repertoire?
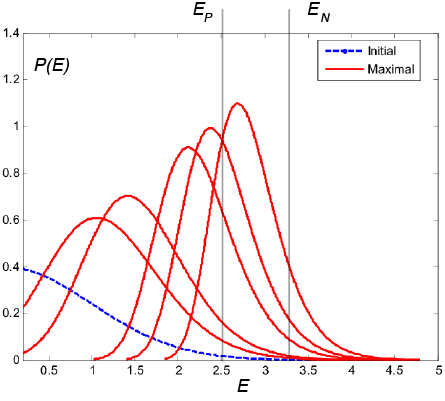
![]() Simulations were performed for TCRs of length N=5 and varying values of M
Simulations were performed for TCRs of length N=5 and varying values of M
(strong binding) 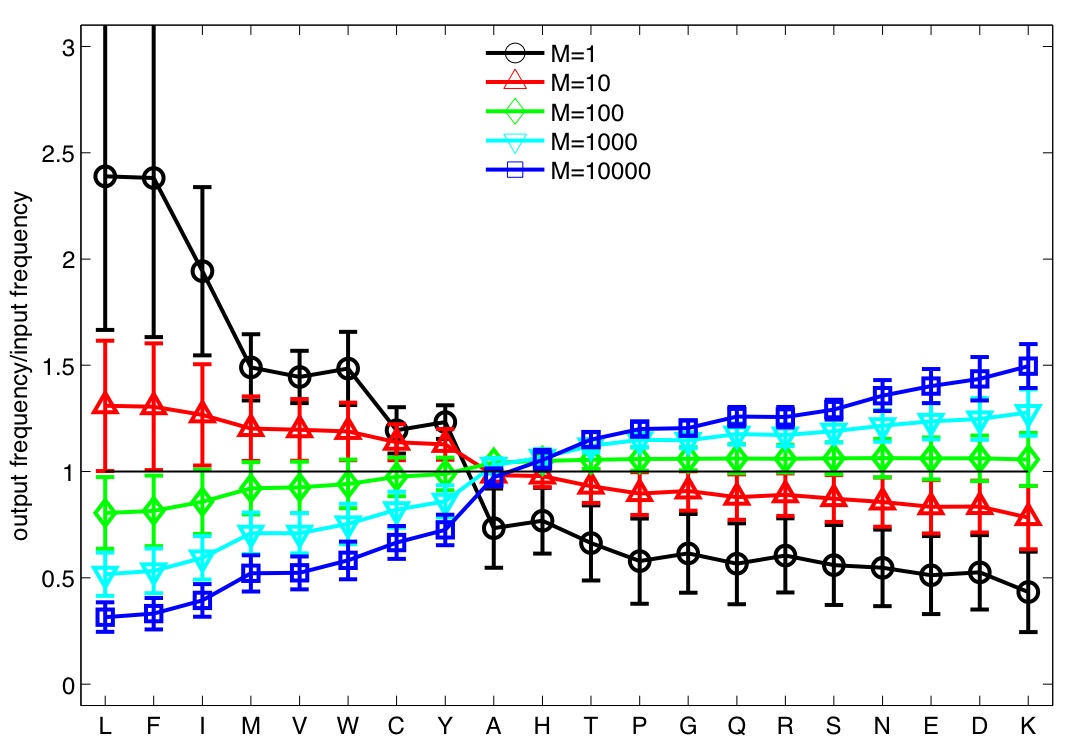 (weak binding)
(weak binding)
(Typical number of peptide encounters in thymus is 1,000-10,000)
![]() Specificity of TCR comes from cooperative binding of several weakly interacting amino-acids (team effort).
Specificity of TCR comes from cooperative binding of several weakly interacting amino-acids (team effort).
Verified in transgenic mice [Huseby, et. al, Nat Immunol 7, 1191 (2006)]
Created mice expressing only one self peptide (M=1) in their thymus!
Compared specificity of TCRs from normal and transgenic mice to mutations of a recognized peptide.
Normal mice TCR typically do not recognize peptides after single site mutations,
while transgenic mice TCR are tolerant of most single site mutations, except at a few "hot spots."
Elite controllers of HIV appear to better tolerate mutations of the virus. Is a similar mechanism at play?
A. Košmrlj, ..., A.K. Chakraborty, Nature online (5 May 2010) WebMed, ...
![]() Connection to Statistical Physics:
Connection to Statistical Physics:
The selection condition is equivalent to the choice of the Extreme Value:
![]()
Characteristics of the Extreme Value Distribution:
Binding energies of a particular TCR sequences:
Extremal mean value:
where
and
are the mean and variance of interactions of the candidate TCR sequence.
Extremal standard deviation:
Note scaling in the large N limit;
(proteome)
Due to the sharpness of the distribution in the large N limit, the deletion condition can be written as:
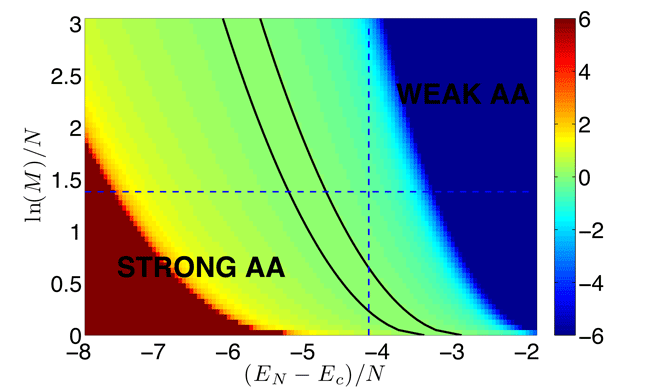
![]() The above selection condition is reminiscent of the micro-canonical rule in Statistical Physics.
The above selection condition is reminiscent of the micro-canonical rule in Statistical Physics.
The "energy" involves interactions amongst the N amino-acids in the sequence
The "interaction" depends only on the sum of variances for the individual amino-acids
As such, in the large N limit, the probability to select a sequence can be written as a product
,
where
and
have to be obtained self-consistently from
and
.
![]()
![]()
Because of the restriction to an energy interval, there is a range of parameters where
= 0.
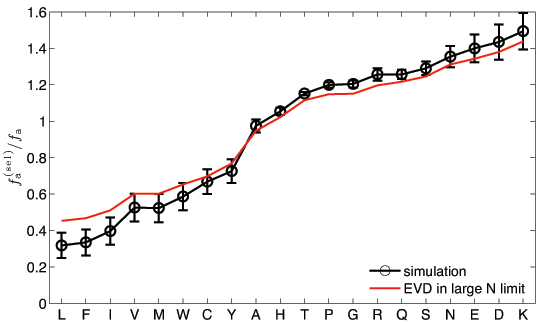
![]() How well does this work for finite N? (N=5 and M=10.000)
How well does this work for finite N? (N=5 and M=10.000)
A. Kosmrlj, A.K. Chakraborty, M.K., & E. Shakhnovich, PRL 103, 068103 (2009)

=E_.jpeg)